Differences in our DNA have major influences on how our genomes function, but how?
Researchers from the NHGRI-funded Impact of Genomic Variation on Function Consortium are exploring how genomic variation affects genome function.
While most DNA letters are the same amongst everyone, just under 0.5% are different between any two people. Over the past two decades, researchers have identified hundreds of millions of these differences, known as genomic variants, that exist across the human population.
Each person has several million genomic variants and only a very small subset of these variants play a role in human health and disease — but which ones?
How genomic variations affect genome function and in turn influence human traits is challenging to study partly because of the sheer number of genomic variants that have been identified.
To understand the functional effects of genomic variation, the National Human Genomic Research Institute (NHGRI), part of NIH, launched a consortium in 2021 called Impact of Genomic Variation on Function (IGVF). The NHGRI-funded consortium now involves researchers from over 120 laboratories across the country who are using multiple experimental and computational techniques to determine the effects of genomic variation.
The IGVF Consortium recently published a paper in Nature outlining the specific goals and approaches they are using to develop a more systematic approach to understanding the impact of genomic variation on function.
“A driving challenge in the genomics field right now is characterizing the rapidly growing catalog of genomic variants with unknown functions,” says Mike Pazin, Ph.D., program director in the Division of Genome Sciences. “Identifying those variants is the easier part, but understanding what they all do is the challenge.”
With more genomic variants being discovered every day, IGVF researchers are developing and testing various model systems and techniques — and using a synergy of experimental and computational approaches — to unravel the complex interplay of genomic variation and genome function. IGVF researchers are working collectively to integrate this information into a catalog of variant impacts, including standards, data, tools and models that will be shared with the broader research community.
A driving challenge in the genomics field right now is characterizing the rapidly growing catalog of genomic variants with unknown functions. Identifying those variants is the easier part, but understanding what they all do is the challenge.
Creating ways to screen for genomic variants related to disease
One of the many approaches that IGVF researchers are taking to understand the impact of genomic variation is to systematically test variants to determine which ones can affect gene expression in common human diseases.
Hyejung Won, Ph.D., an associate professor of genetics at the University of North Carolina (UNC) at Chapel Hill, is investigating regulatory elements — regions of the genome that can dampen or amplify the expression of genes, similar to how a dial on a radio turns music louder or softer.
Using an efficient technique called massively parallel reporter assay, Dr. Won and her research group, which includes her UNC colleagues Karen Mohlke, Ph.D. and Michael Love, Ph.D., are planning to screen around 250,000 variants for their function in regulating other genes and in various organs in mice. They also plan to understand these variants in female versus male mice and in mice with and without inflammation.
The specific variants being studied were previously found to be associated with health conditions such as brain disorders, metabolic disorders, respiratory disorders, cardiovascular diseases and muscular disorders.
The results generated by these studies will be validated by other IGVF researchers using different methods. IGVF researchers will also use the findings to develop predictive models of unstudied variants and how these variants behave in different cell types or environments. Collectively, this work will show which genes are controlled by these regulatory elements, whether variants in the regulatory elements increase or decrease gene activity and how the variants are affecting traits in the cells, organs and the body.
“IGVF researchers are working in multiple groups to address different aspects of the workflow,” says Dr. Won. “My group’s role is to understand: What is the variant’s function? What variants would we like to focus on? We hope that by working with other IGVF researchers, we will understand how these variants affect human health and disease.”
Developing a network-level understanding of variation: Using insulin production as an exemplar system
IGVF researchers are also examining how genes and regulatory elements may be forming networks of interrelated parts and how genomic variation behave in specific cell types and phenotypes.
Hannah Carter, Ph.D., an associate professor at the University of California, San Diego, is part of a research group taking a network-level approach focusing on specialized cells in the pancreas called beta cells.
Beta cells make insulin to help convert sugar in the blood to an energy source for the body. In people with diabetes, the body stops making and/or stops responding to insulin, causing high blood sugar levels. Over time, high blood sugar levels can damage organs and lead to complications such as strokes, heart attacks and kidney damage.
Dr. Carter’s group studies a beta cell model not only because beta cells are relevant to diabetes but also because researchers can observe the dynamic nature of insulin production.
“The changes in insulin levels are on a short timescale that is easy to study,” says Dr. Carter. “It’s a tightly regulated process that can be affected by a number of different factors, from genomic variation to diet to cellular stress.”
A collaborative group of UC San Diego colleagues that includes Maike Sander, M.D., Bing Ren, Ph.D. and Kyle Gaulton, Ph.D., are using pancreatic cells in tiny, three-dimensional tissue cultures, known as organoids, to study how genomic variants in different contexts can impact how much insulin is produced. To simulate different environmental conditions such as inflammation and cellular stress, the researchers expose the organoids to different chemicals or small proteins and then observe the effects. Together, the researchers are working on ways to identify a network of interrelated parts of the genome that fine-tune insulin levels in the body.
“In addition to glucose, there are different factors that affect insulin production that are not well understood,” said Dr. Carter. “It’s important to study how genomic variants, genetic background and environmental factors all tie in together to accurately predict risk and come up with effective solutions to help control the disease. It’s the missing piece in the diabetes puzzle.”
The work being done by Dr. Carter and colleagues, along projects by other IGVF researchers using different cell types and phenotype models, aim to inform optimal strategies that can be applied to other biological systems.
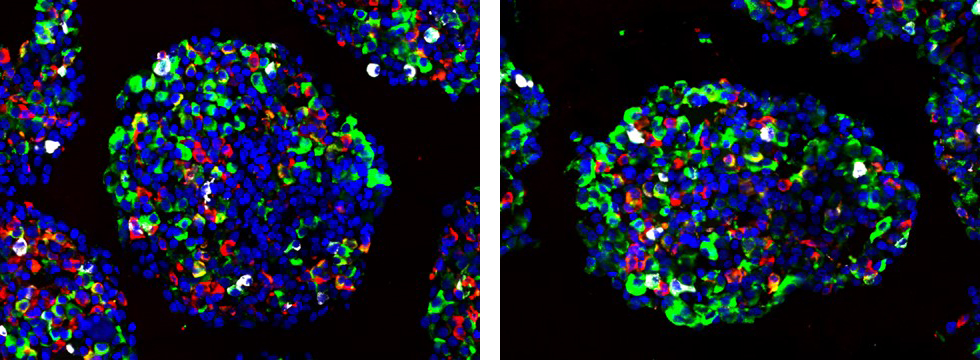
Caption: Microscope images of an organoids that models pancreatic islets. Pancreatic islets are a group of cells in the pancreas that produce hormones to control blood sugar levels. The hormones include glucagon (green), insulin (red), somatostatin (white). The nuclei of the cells are blue. Image credit: Kim-Vy Nguyen-Ngoc, Ph.D., University of California, San Diego.
Incorporating computational approaches to characterize variants
IGVF researchers are using both experimental and data science approaches to tackle challenges in characterizing genetic variants. This includes using new and emerging computational methods, approaches and tools to decide which variants to test. They are also finding new ways improve the sensitivity and scale of functional assays and to develop predictive models.
Luca Pinello, Ph.D., an associate investigator at the Massachusetts General Hospital, is combining genome-editing tools and computational methods to study genomic variation. Working with Daniel Bauer, Ph.D. (Boston Children’s Hospital), Guillaume Lettre, Ph.D. (Montreal Heart Institute) and Richard Sherwood, Ph.D. (Brigham and Women’s Hospital), he is seeking to understand how variants within genomic regulatory elements affect cardiovascular diseases and blood traits.
According to Dr. Pinello, heart and blood disease traits are amongst the most studied traits in large-scale genomic studies. These traits have been linked to numerous genomic variants that have yet to be assessed for their effect on cells and the body.
The researchers are developing innovative computational tools to select which genomic variants to study first. Then, they will use technologies based on the gene editing system CRISPR to add or change variants in the genomes and observe how the variants affect cellular traits.
The researchers have already made progress in developing a new method called BEAN (Base Editor screen analysis with Activity Normalization), which allows genomic variants to be more accurately identified. BEAN refines an existing approach called base editing screening, in which researchers create and study the functions of large numbers of genomic variants. BEAN has already helped the researchers discover genomic variants that affect cholesterol uptake in liver cells. The researchers are also working to optimize CRISPR gene editing techniques in an effort to create new variants and predict their influence on genome function.
As is the case for all IGVF studies, the results of this research will contribute to the larger IGVF catalog of variant impacts. The scientific community can then use this catalog to inform future research on different disorders and diseases.
“We believe that the most groundbreaking discoveries happen at the intersection of different fields,” says Dr. Pinello. “We're pushing the boundaries of what's possible in understanding genomic variation, thanks in part to the diverse expertise across the different centers within the IGVF Consortium.”
Last updated: September 5, 2024
