Three new approaches tackle challenging design problems in genomics technology development
Researchers funded by NHGRI’s Genome Technology Program include a wide range of investigators with a variety of scientific expertise.
The advent of new DNA sequencing techniques has been a gamechanger in genomics. However, new genomic methods and technologies are still needed to advance basic and clinical research investigating human biology and disease. To address this challenge, the National Human Genome Research Institute (NHGRI), part of NIH, supports research to innovate and develop novel approaches and technologies through the Genome Technology Program.
Over the past 20 years, the program has fueled the development of novel technologies for studying the human genome, including deepening our understanding of genomic variants and their functions. Initially focused on advancing nucleic acid sequencing, the program has since expanded and now supports the development of additional genomic technologies. These technologies help scientists advance key areas such as: understanding gene regulation and genome function; synthesizing nucleic acids; and examining nucleic acids and their modifications at a single-molecule level.
Innovation in genomic technologies drive the field forward, allowing researchers to understand fundamentals of biology and find new ways to improve human health. By supporting researchers in their pursuit of new and creative ways to address scientific challenges, we hope these innovations will continue to contribute to new genomic discoveries and knowledge.
The program also helps researchers disseminate their technologies and approaches to the broader research community, which includes helping to put new methods on the path towards commercialization.
With a renewed interest in understanding RNA and delving deeper into its biology, including a focus on RNA sequence and modifications, here are snippets of some exciting RNA-focused projects that are supported by NHGRI’s Genome Technology Program.
Improving accuracy of DNA and RNA sequencing
Recent advances in nucleic acid sequencing methods help researchers analyze long nucleic acid molecules (1,000 nucleotides or more in length), a technique known as long-read sequencing.
One type of long-read sequencing, called nanopore sequencing, involves threading strands of DNA or RNA nucleotides through small pores within a membrane, generating changes in electric currents that are distinct for each type of nucleotide. While the technique has high accuracy for DNA sequencing, directly reading RNA nucleotides has proven to be tricky due to the modifications that exist on RNA molecules.
Aleksei Aksimentiev, Ph.D., a professor at University of Illinois at Urbana-Champaign, leads a group that aims to improve the accuracy of nanopore sequencing using computational approaches.
“We know how nanopore sequencing works, but we don’t know why,” he says. “Why are the currents what they are? There is a fundamental knowledge gap in linking sequences to currents.”
A physicist by training, Dr. Aksimentiev considers this question one of the most interesting (and most physics-intensive) problem in biophysics. Collaborating with Min Chen, Ph.D., a professor at the University of Massachusetts Amherst, Dr. Aksimentiev and colleagues are developing computational technologies that will help researchers custom design nanopores to more accurately detect nucleotides. They hope to take their improvements a step further by helping researchers more accurately detect modifications that exist on RNA molecules, an area of much interest as described by a recent report from the National Academies of Sciences, Engineering and Medicine.
Dr. Aksimentiev and his group are using a method called all-atom-molecular dynamics, a computer simulation technique that analyzes how atoms move and interact. Through this method, the researchers are working towards understanding how the different changes occur at a nanoscale level.
Eventually, they hope to use the results from their research to develop software that better allows researchers to decode DNA or RNA sequence and that can be used to better design nanopores to detect nucleotides more accurately.
“It's our goal to make our technology accessible, transferable and usable to others,” he says.
Related Content
Finding and analyzing RNA modifications by developing standards
Though RNA modifications are thought to influence human health and disease, our understanding of them has been limited by methods and technologies that are currently available.
Sara Rouhanifard, Ph.D., a professor at Northeastern University, is hoping to change that. She collaborates with a team that includes Meni Wanunu, Ph.D. (Northeastern University) and Ya-Ming Hou, Ph.D. (Thomas Jefferson University), to help researchers more efficiently and accurately detect RNA modifications. Specifically, the group has developed a method to identify RNA modifications and, more recently, to create standards (or controls) to quantify them.
“RNA modifications are extremely prevalent in transcriptomes, and if the cells are putting in so much energy to make these modifications, there has to be a biological reason,” she says. “We’re interested in finding out what that reason is.”
Researchers can use direct RNA nanopore sequencing to detect RNA modifications, but it can be difficult to interpret many of the signals that are associated with modifications without known standards to use for comparison and calibration.
To address this, Dr. Rouhanifard and her team are developing standards of synthetic RNAs with known modifications as well as standards that can be used to assess baseline signals, so that researchers will know what each modification’s signal looks like and how to interpret the resulting data. In particular, they are working to develop standards for pseudouridine modifications, a common type of modification on mRNA molecules. While pseudouridine modifications have the potential to change the structure and function of the mRNA molecule, the exact effects of the modifications are difficult to establish.
Similar to how mushroom foragers have a guidebook to know what a specific type of mushroom looks like, having standards for RNA modifications allow researchers to interpret a given signal and determine which RNA modification is causing it. Without an example that includes details of what to look for, it can be difficult to know exactly which RNA modification is being detected.
Developing RNA standards is challenging because RNA molecules are unstable and synthesizing RNA strands with precise modifications can be costly. Because of these constraints, Dr. Rouhanifard’s group has been taking a more laborious approach of piecing together smaller strands of RNA with known modifications to guarantee the purity of the final product.
So far, this approach of making and testing standards has helped the researchers detect new sites on human mRNA containing pseudouridine modifications and define sites that respond dynamically to changes in cellular state, setting the stage for other researchers to properly find and characterize RNA with pseudouridine modifications.
"The primary challenge with using nanopore sequencing to identify RNA modifications is separating the signal from the noise, and the only way to do that is by generating and using appropriate standards,” says Dr. Rouhanifard.
Looking at mRNAs of a single cell in its natural environment
In addition to accurately reading and analyzing RNA sequences, being able to capture RNA molecules in the first place is crucial for addressing fundamental questions in biology.
Many of the current approaches to investigating mRNA have to choose between looking at the expression of a few select genes in a whole tissue or looking at more genes in cells grown in an artificial environment. While the first approach allows cells to behave in their natural environment, the second approach provides a more comprehensive look at gene expression.
For Fei Chen, Ph.D., professor at the Broad Institute at MIT and Harvard, the question is, “Why not both?”
Cells are known to communicate and coordinate with neighboring cells in order for tissues, organs and the whole body to function in concert. Capturing mRNA in whole tissues allows researchers to have a more complete understanding of how all genes turn on and off.
Recently, Dr. Chen collaborated with Evan Macosko, M.D., Ph.D., to build a genomics platform to comprehensively study gene activity of cells within intact tissues — without removing the cells from their natural environment. Their goal was to use a new tool, called Slide-Tag, to contextualize single-cell gene expression and understand genome function at the tissue level.
To do this, the researchers built an array of beads that have unique barcodes that can be used to detect their location. The beads are small, around the size of a single cell. Researchers can put sample tissues on the array, with each bead helping them to locate individual cells within the tissues (akin to GPS for the cells).
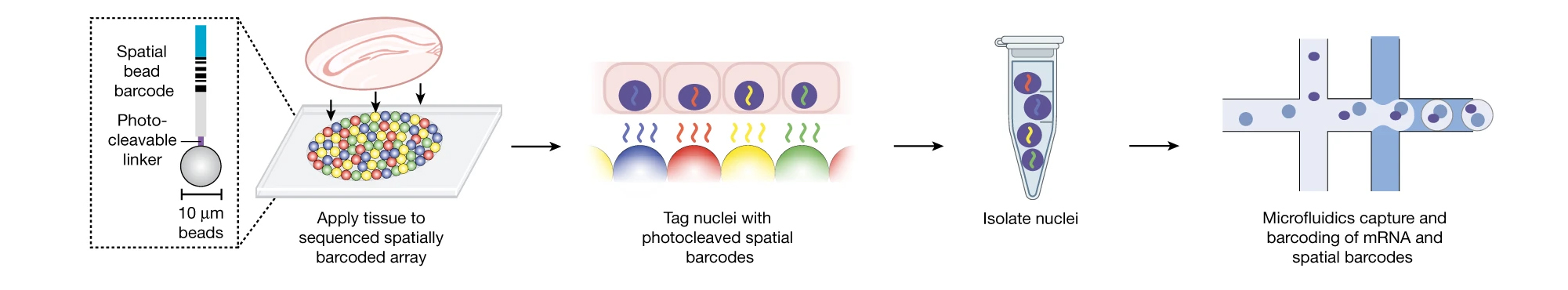
“The array of beads is like the group of pixels in a camera picture. Instead of taking a photo, we can capture gene expression of many genes in the context of the whole tissue,” says Dr. Chen.
Already, the group has applied their technology, and a similar previous technology, Slide-seq, to probe intricate cellular details of the mouse brain and how immune cells change in different environments in a tumor. With further developments, Dr. Chen hopes that other researchers can start using the tool to understand complex biological processes such as development and cancer.
Supporting the development of novel genomic technologies
While these snippets offer a glimpse into the world of genome technology development focused on RNA, many more projects are underway. Recent publications coming out of NHGRI’s program are highlighted in videos created by the researchers themselves.
NHGRI has also established a central hub that connects funded researchers working on technology development, known as the NHGRI Technology Development Coordinating Center. This center organizes regular events, communicates about program news and develops resources to share with the broader research community.
The NHGRI Genome Technology Program also works in close collaboration with NHGRI’s Small Business Program to encourage researchers to develop innovative genomic technologies in the private sector that have a strong potential for commercialization.
“Innovation in genomic technologies drives the field forward, empowering researchers to understand fundamentals of biology and find new ways to improve human health,” says Stephanie Morris, Ph.D., Lead Program Director for the NHGRI Genome Technology Program. “By supporting researchers in their pursuit of developing new and creative ways to address scientific challenges, we hope these innovations will yield new genomic discoveries and knowledge.”
NHGRI’s Technology Development Coordinating Center is hosting an event on June 10, 2024, that will be open to the public. The Genome Technology Forum will bring together industry leaders, genome technology developers and genomics researchers to showcase how these new technologies can be used and to discuss the opportunities and challenges associated with their optimization and use. More information and registration can be found here.
Last updated: May 29, 2024